Medical imaging is pivotal in modern healthcare, enabling diagnosis, treatment planning, and disease monitoring across modalities like MRI, CT, and pathology slides. However, developing robust AI models for these complex and high-dimensional datasets demands specialized tools that go beyond general-purpose Deep Learning frameworks.
Enter MONAI (Medical Open Network for AI), a PyTorch-based, open-source ecosystem crafted explicitly to accelerate and simplify Deep Learning in Medical Imaging. MONAI combines domain-specific data handling, advanced neural architectures, and optimized workflows to empower researchers and clinicians alike.
- Why MONAI? Tailored for Medical Imaging on the PyTorch Ecosystem
- Core Features of MONAI
- Advanced Capabilities Enhancing MONAI Power
- Performance and Scalability: GPU Acceleration and Distributed Training
- C++/CUDA Optimized Modules in MONAI for Domain-Specific Routines
- MONAI Bundles: Portable, Reproducible Model Packages
- Conclusion
- References
Why MONAI? Tailored for Medical Imaging on the PyTorch Ecosystem
While PyTorch provides a flexible base for deep learning, MONAI extends this foundation with medical imaging-specific capabilities. Key motivations include:
- Collaborative Development: A community-driven platform uniting academic, industrial, and clinical researchers on shared tools.
- End-to-End Training Workflows: Ready-to-use, state-of-the-art pipelines optimized for medical imaging tasks.
- Standardized Model Evaluation: Consistent methodologies for creating, training, and evaluating deep learning models tailored for healthcare.
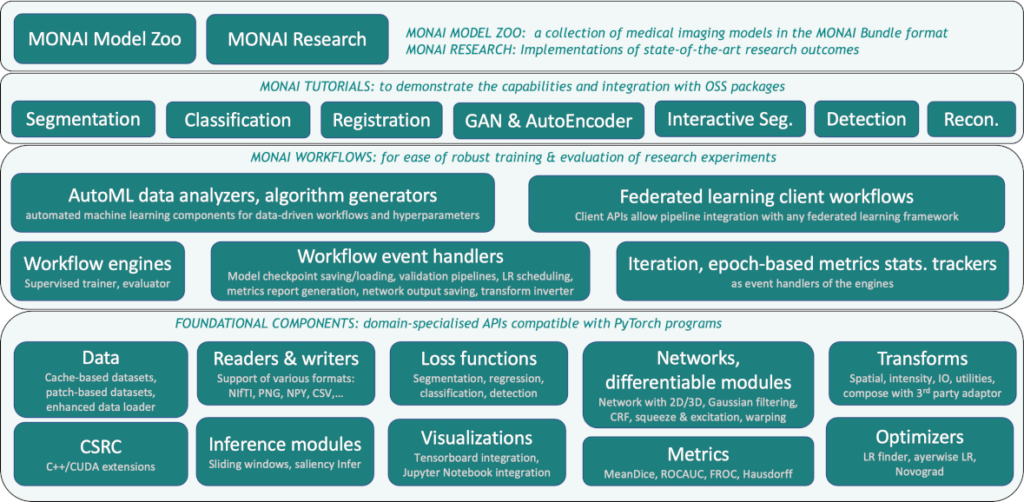
MONAI’s layered architecture builds on Python and PyTorch, adding specialized bundles, labelers, and deployment tools, ensuring seamless integration and extensibility.
Core Features of MONAI
Flexible, Domain-Specific Data Handling and Augmentation
Medical images require handling complex formats (e.g., DICOM, NIfTI) and multi-dimensional data arrays. MONAI’s monai.data and monai.transforms modules provide:
- Preprocessing pipelines for 2D/3D/4D data.
- Transformations in both array and dictionary styles, enabling synchronized augmentation of images and labels, which is essential for segmentation and multi-modal tasks.
- Advanced patch-based sampling with weighted and class-balanced strategies to address imbalanced datasets, crucial in medical imaging.
These capabilities surpass typical PyTorch data loaders by addressing domain-specific needs such as metadata handling and spatial consistency.
Pre-Built Networks, Loss Functions, and Optimizers for Medical Tasks
MONAI implements neural architectures designed to process spatial medical data (1D, 2D, and 3D), with utilities to fine-tune pretrained weights from sources like MMAR or the MONAI Model Zoo.
In contrast to general PyTorch implementations, MONAI includes medical imaging-centric loss functions such as:
- DiceLoss and GeneralizedDiceLoss for segmentation overlap.
- TverskyLoss loss to control false positive/negative trade-offs.
- DiceFocalLoss combines class imbalance handling and hard-example mining.
Optimizers like Novograd and utilities such as LearningRateFinder help tailor training to the unique properties of medical datasets.
Evaluation: Sliding Window Inference and Medical Metrics
Processing large 3D volumes often exceeds GPU memory constraints. MONAI’s sliding window inference efficiently processes sub-volumes sequentially, supporting overlapping windows and blending for smooth predictions.
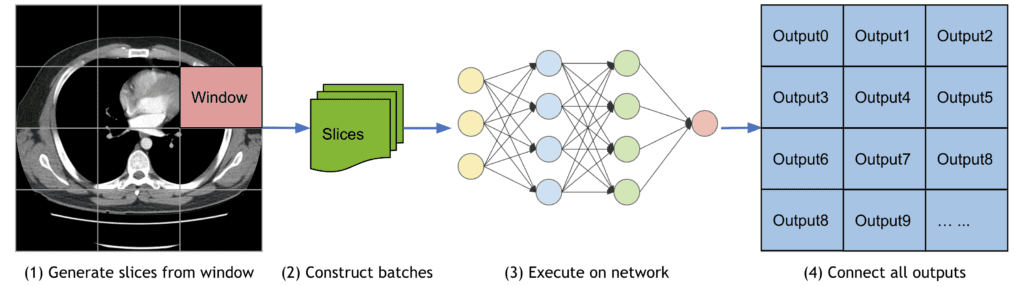
Extensive metrics cater specifically to medical imaging evaluation:
| Metric | Purpose |
|---|---|
| Mean Dice | Segmentation overlap accuracy |
| ROCAUC | Classification performance |
| Confusion Matrices | Detailed classification outcomes |
| Hausdorff Distance | Shape boundary similarity |
| Surface Distance | Average boundary distance |
| Occlusion Sensitivity | Model robustness testing |
Additionally, MetricsSaver facilitates comprehensive report generation with statistics like mean, median, percentiles, and standard deviation.
Visualization Tools for Insightful Data and Model Interpretation
Beyond typical plotting, MONAI integrates with TensorBoard and MLFlow to visualize:
- Volumetric inputs as GIF animations.
- Segmentation maps.
- Intermediate feature maps.
The utility matshow3d offers slice-by-slice visualization of 3D images using matplotlib, aiding in qualitative assessment.
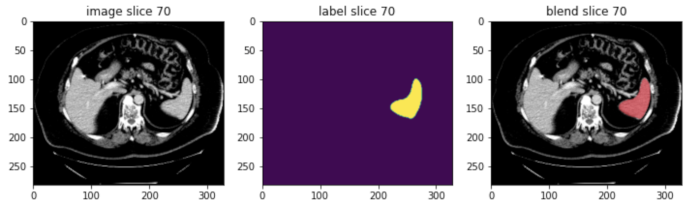
Moreover, MONAI’s blend_images function overlays segmentation labels on images, enhancing interpretability for clinical review, a feature not standard in base PyTorch workflows.
Modular Workflows and Event Handlers
MONAI’s training and evaluation are structured via PyTorch Ignite engines and event handlers, offering:
- Clear decoupling between domain-specific logic and generic machine learning operations.
- High-level APIs supporting AutoML and federated learning.
- Event-driven control enabling automatic metric logging, checkpointing, learning rate scheduling, and validation.
This modular approach streamlines reproducibility and customization, simplifying complex experiments.
Advanced Capabilities Enhancing MONAI Power
Ensemble Learning with EnsembleEvaluator
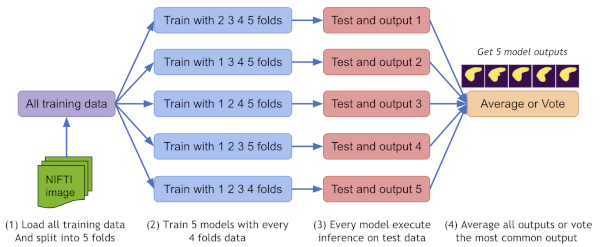
MONAI supports cross-validation-based ensembling by splitting datasets into K folds, training K models, and aggregating predictions via averaging or voting. This enhances model robustness and generalization, vital for sensitive medical applications.
Auto3dseg: Automated Large-Scale 3D Segmentation
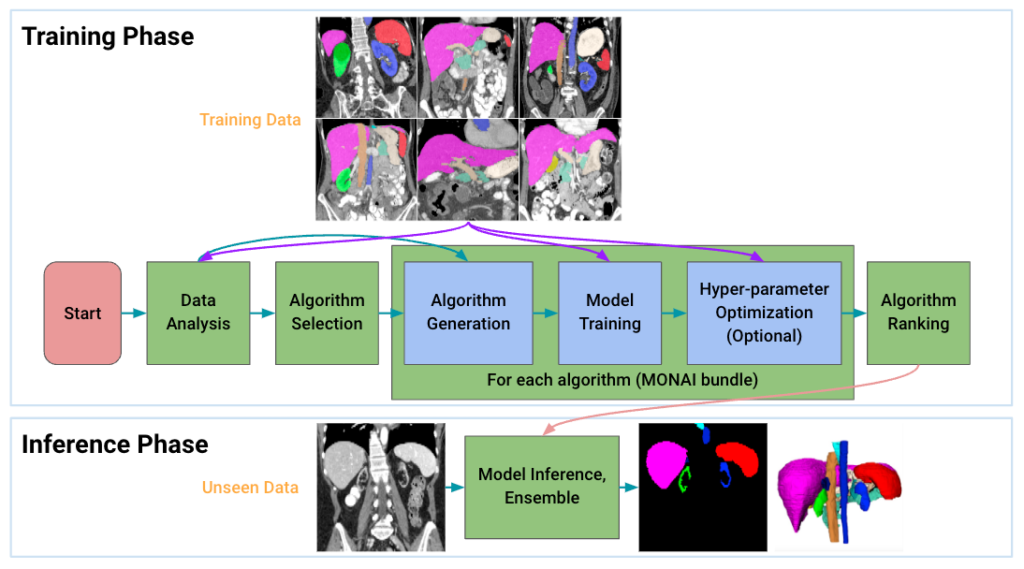
Auto3dseg automates the entire segmentation workflow by:
- Analyzing data statistics globally.
- Generating MONAI bundle algorithms dynamically.
- Training and hyperparameter tuning.
- Selecting top algorithms via ranking.
- Producing ensemble predictions.
This solution bridges beginner-friendly usage and advanced research needs, validated on diverse large 3D datasets.
Performance and Scalability: GPU Acceleration and Distributed Training
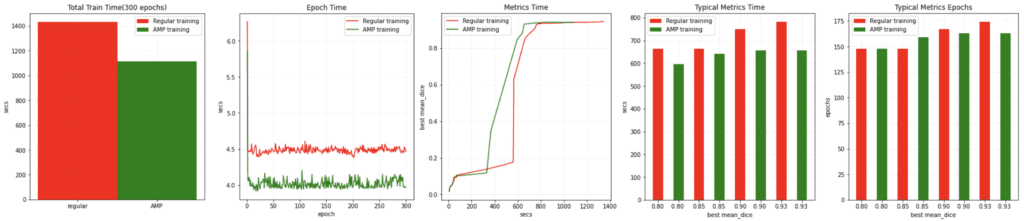
Auto Mixed Precision (AMP)
AMP training in MONAI leverages NVIDIA’s hardware capabilities to reduce memory and speed up training with minimal accuracy compromise. Benchmarks on V100 and A100 GPUs show a significant reduction in training times and faster metric computations compared to non-AMP training.
Profiling Tools
Integration with NVIDIA tools like DLProf, Nsight, NVTX, and NVML allows fine-grained performance analysis to identify bottlenecks and optimize workflows.
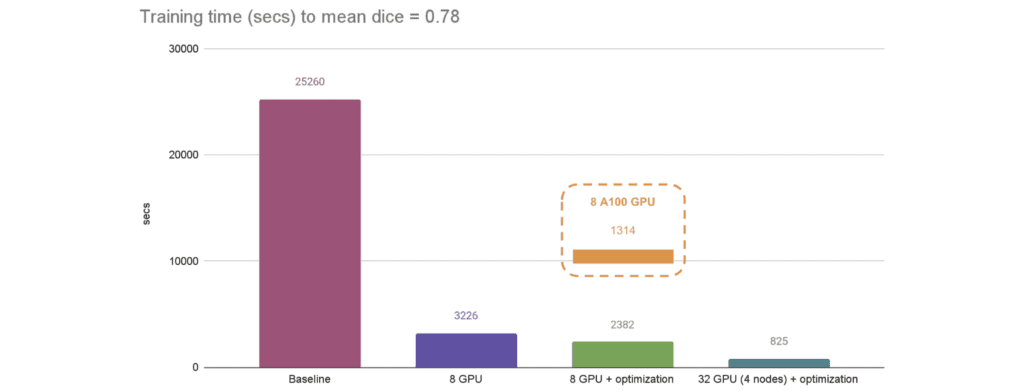
Distributed Training
MONAI’s APIs align with PyTorch’s native distributed module, Horovod, XLA, and SLURM. Distributed training scales efficiently across GPUs and nodes, with demonstrated speedups from single-GPU baselines to 32-GPU multi-node clusters. Combined with AMP, caching datasets, and optimized loaders, this ensures rapid model development on large datasets.
C++/CUDA Optimized Modules in MONAI for Domain-Specific Routines
To maximize performance in critical steps, MONAI incorporates C++/CUDA extensions for operations such as:
- Resampling – refers to changing the spatial resolution or dimensions of medical images to a standardized or desired size.
- Conditional Random Fields (CRF) – a statistical modeling approach used primarily as a post-processing method to refine segmentation predictions. CRFs consider contextual spatial relationships among pixels or voxels, smoothing segmentation results, and correcting localized inconsistencies.
- Fast Bilateral Filtering – a non-linear edge-preserving smoothing technique. Unlike standard filtering that blurs all image details equally, bilateral filtering smoothes images while preserving sharp edges based on intensity and spatial proximity.
- Gaussian Mixture Models for Segmentation – assume image intensities or feature spaces can be modeled as a combination (mixture) of several Gaussian distributions. Pixels are segmented into different tissue classes based on their statistical characteristics, represented by these Gaussian distributions.
These modules accelerate workflows beyond standard PyTorch implementations.
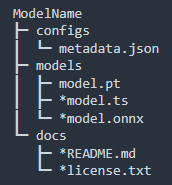
MONAI Bundles: Portable, Reproducible Model Packages
Bundles encapsulate models with weights (PyTorch, TorchScript, ONNX), metadata, transform sequences, legal info, and documentation in a self-contained package. This enables easy sharing, deployment, and reconstruction of training/inference workflows, promoting reproducibility and usability in clinical and research environments.
Conclusion
MONAI stands out as a specialized, extensible, and high-performance framework built on PyTorch, explicitly addressing the challenges of medical imaging AI. From optimized data pipelines and pretrained model repositories to advanced workflows, visualization, and distributed training, MONAI empowers researchers and clinicians to accelerate innovation and deployment in healthcare.
Whether someone is a beginner seeking turnkey solutions like Auto3dseg or an expert customizing pipelines with event handlers and C++ extensions, MONAI offers the tools and community support to advance medical AI reliably and efficiently.




100K+ Learners
Join Free OpenCV Bootcamp3 Hours of Learning